The two main questions in most microbiome projects are
- What “little bugs” are in the sample?
- What is the metabolic potential of the community?
Maybe you feel nervous about the descriptive nature of the questions; it is usually more interesting to explain processes rather than static components. However, once you can determinate the taxonomic and genetic profiles of your samples, you can start to make corelations of these profiles based on time or environmental variables.
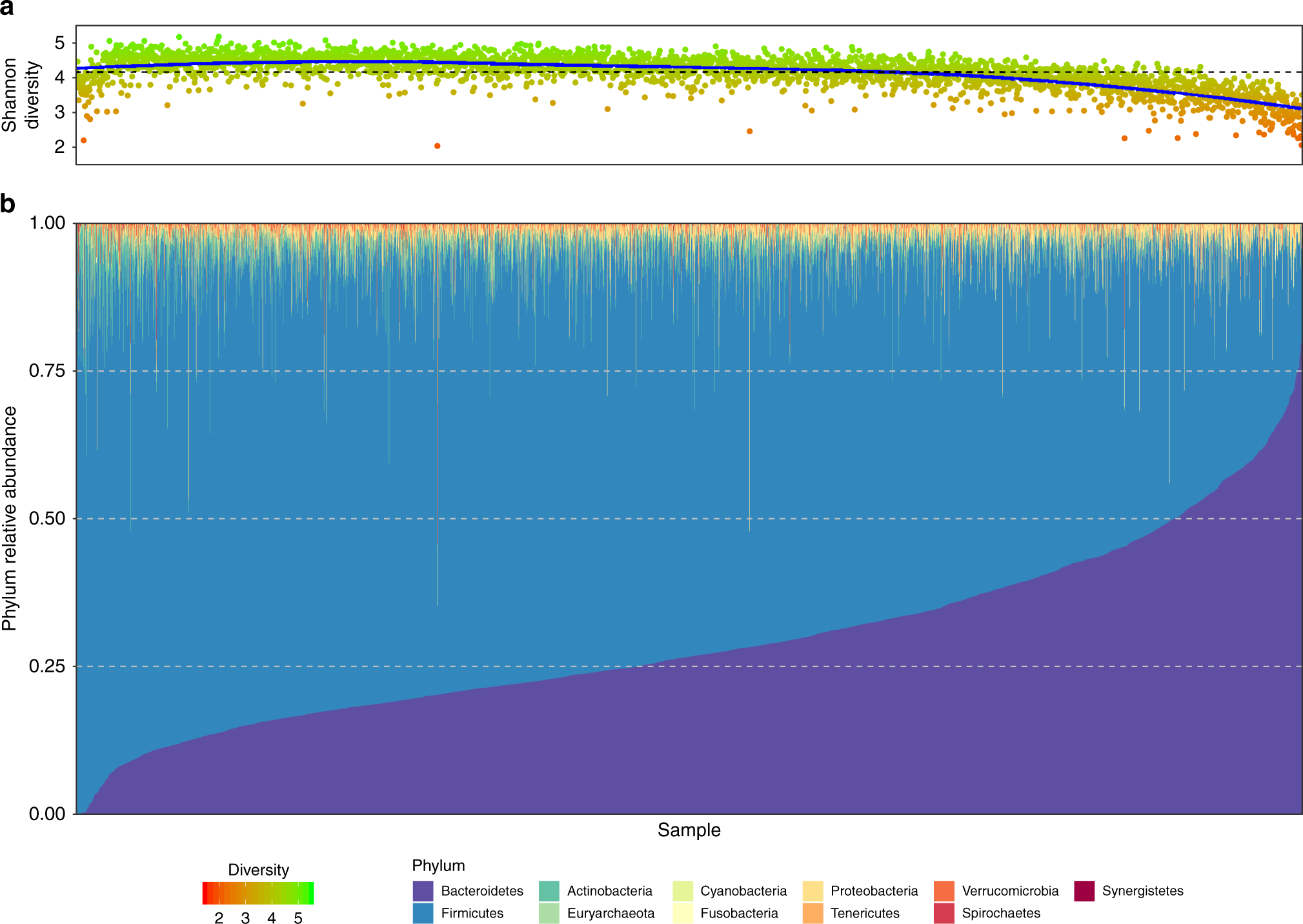
The microbiota has showed convergence and variability at the same time; there are core species in samples belonging to the same environment, but there are also specific microbes for each sample. The corelation studies allow us to overcome the large amount of data and detect the most evident core/specific microbes. The results of these studies range from the proposition of microbes as pathogens (due to their absence in healthy plants, and presence in sick ones), to the development of diagnostic tools. For example, the Knight lab has a machine learning algorithm capable of detecting X cancer based in a blood microbiome sample (the taxonomic profile) :O.
If you are still interested in more than corelations, let’s talk about (https://www.nature.com/articles/s41467-020-17956-1). The study is interested in the taxonomic and genetic profiles. Specifically in microbiomes of Crohn’ disease patients (CDP), and the microbiome of patients after treatment (RP) at different stages of the treatment, and outcomes of the treatment (remission or relapse). Nevertheless, the research group also wants to prove that the composition and diversity of the microbiome in mouse models change the disease (Crohn’ disease) state of the host. The new goal can seem like a subtle distinction with respect to the corelation studies, but it demands an experimental model.
The corelation between group of patients and taxonomic profiles show that the diversity indexes of the microbiome change based in the stage of treatment, state of the disease, etc. Besides, the study identifies differential abundance species across groups of patients. On the other hand, the experimental model show that the CD clinical picture can be induced in germ-free mice after CD patients’ fecal transplantation; the taxonomic composition can induce Crohn’ disease. Finally, the study makes correlational studies between the taxonomic and genetic profile in the samples, a common strategy to create hypothesis of the host-microbiome interactions. This work underlines the relevance of the identity and abundance of the microbes in the phenotype, with corelation studies and an experimental model!
Once pointed the importance of the taxonomic profiles in the microbiota and the diversity of approaches to estimate these profiles, I will talk about the common strategies of taxonomic profiling. Some gold standards have been proposed, however, go out of the recipe can improve you study with the correct experimental design.