THE ABC OF SERVERS AND CLUSTERS
If you work in bioinformatics, you may have to process large datasets with complex scripts. Probably, your computer does not have enough resources for it, you would have to use servers/ clusters :O. Based in my experience, I will recover some useful concepts. However, do not hesitate to contact with the cluster/server manager if you have doubts.
Summary…
- Server: System (e.g., a computer) that shares resources/services with a client.
- Cluster: Network of interconnected computers/servers that coordinates to perform tasks.
- Formed by nodes.
- Types of nodes:
- Head node: Interacts with users and is used to submit jobs, edit scripts, etc.
- Task node: Performs the processes submitted via jobs.
- Types of nodes:
- Formed by nodes.
- Jobs: Processes submitted by users to the servers/cluster (such as execute kaiju).
- A process is a program in execution
- It can have multiple tasks.
- A process is a program in execution
- Advantages of clusters and servers:
- Access to resources
- Centralized access to resources = easier
- Remote access
- Necessary resources for a script execution:
- Memory
- Temporal and used by the programs
- HDD/SDD. Used for long term files.
- Processors/CPU: The brain of the computer
- Formed by cores
- The cores can have threads
- Multiple cores/threads parallelize the tasks of a processes and improves the speed of the processes. But demand larger amounts of RAM.
- Formed by cores
- Memory
Some details…
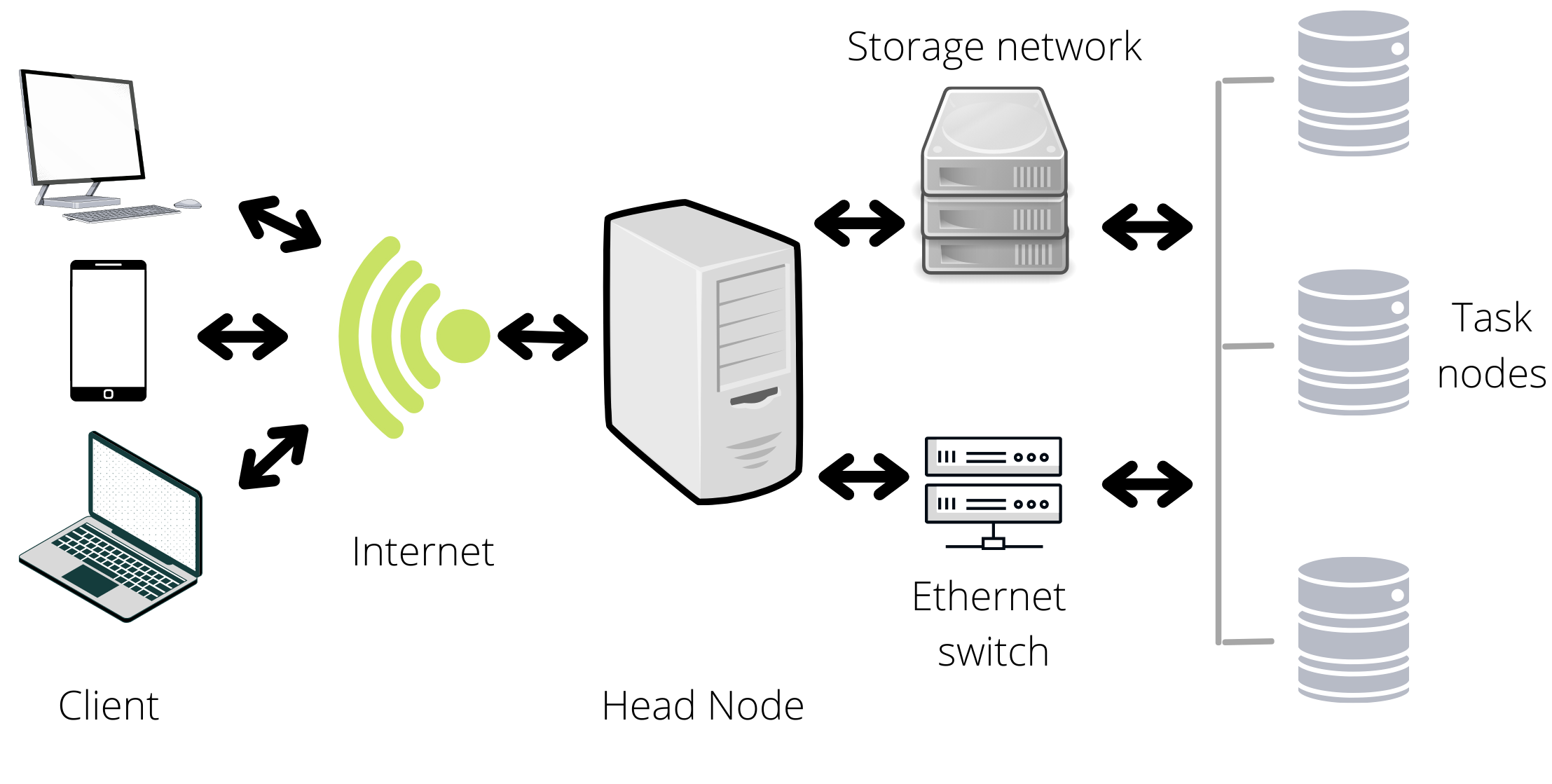
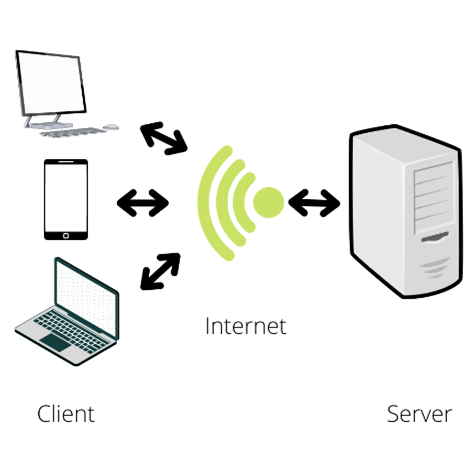
First, let’s define server! A server is a system (e.g., a computer) that shares resources/services with a client (e.g., a university or a person). Some examples of resources/services are storage space, host websites, provide apps, etc. With a server, you can access to large amounts of resources from a remote terminal (your cell phone, laptop, etc.) via internet. The Jobs are defined as processes assigned to the servers by the clients, the jobs are the way we interact with the servers, they can be as simple as print the date in the terminal, or as complex as update the operating system. In complex processes, we can find multiple tasks; for updating the operating system, the computer must download the new files, and print a report in the user terminal, it may have to print the date; in this case a task is a process example. In bioinformatics labs, I have seen the use of servers to storage data bases or data, run scripts and host web sites.
On the other hand, a cluster is a network of interconnected computers/servers that coordinates to perform tasks. Each computer in a cluster is defined as a node (due to graph theory) and there are two main kinds of nodes, the head, and the task nodes. The head node is the one that interacts with the client, this is used to write scripts, or coordinate the submitted jobs. The task nodes are the ones that executes the jobs. The head node should not be used to perform tasks, in other words, you should submit your jobs to the head node so that the head node can distribute it to the task nodes. As a comment, the head and task nodes where previously named master and slave node, but the Black Lives Matter movement has led to a re-examination of the labels.
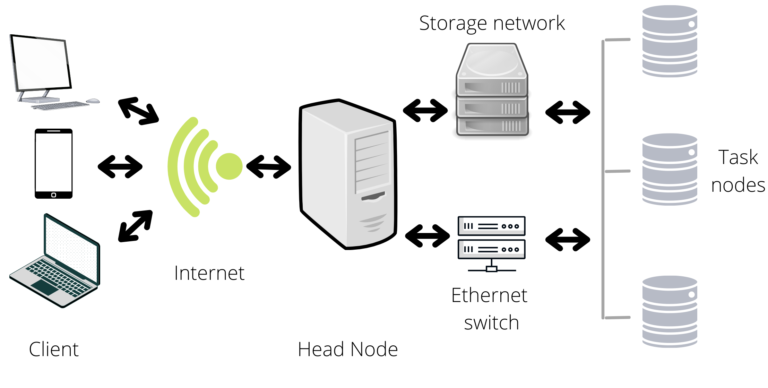
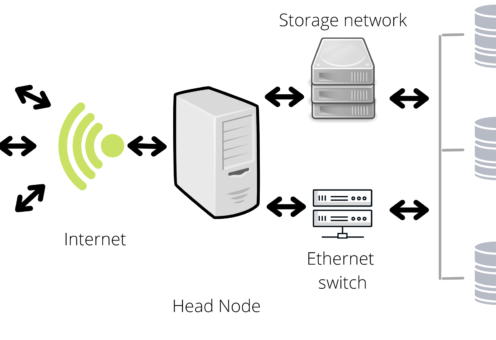
You can find whole books about the functioning of clusters and servers, but three properties will make you love them:
- Since they gather multiple computers, the computational resources increase.
- Due to the interconnected structure, you can organize the resources and script execution from a centralized platform.
- They function in remote access; therefore, you may be able use the clusters/servers from distinct locations/devises.
For example, if you have a microbiome dataset and you want to make taxonomy classifications from them with Kaiju (https://github.com/bioinformatics-centre/kaiju). To do it, you would need enough space to storage the reads (e.g., X MB), the database (e.g., the nr database of Kaiju requires 118 GB), etc.; enough RAM to process the taxonomy classifier, for example, Kaiju uses 200 GB of RAM for the database construction. An average computer has ~ 250GB – 750GB of HDD, 4GB – 16 GB of RAM, and 2-4 cores. Besides, with a server/cluster you may run the analysis in a coffee at the other side of the country.
Now, let’s talk about some computational resources needed for a successful script execution! The memory is the first one, which is used to storage information. The most popular memories are HDD/SSD and RAM. The HDD (Hard disk drive) and the SSD (Solid State Drive) are non-volatile memories, such memories are used to storage long term files since they do not need a constant flux of energy to maintain data. In the microbiome example, you would storage the datasets, the results, and the kaiju scripts in HDDs.
On the other hand, the RAM (Random Access Memory) is a volatile memory, it needs constant energy to retain data; each time you turn off the computer, the RAM is deleted. Nevertheless, it is easier and quicker to access, therefore, you would use it to retain temporal files created by the programs. In the microbiome example, Kaiju uses the RAM to remember the translated protein sequences of the nucleotide input sequences (an intermediate step).
The second resource is the processor, or the CPU (Central Processing Unit), which is referred as the brain of the computers, this one reads/executes the instructions to get a result or redirects the tasks to a specialized hardware. The processing units of the CPUs are the cores, the threads allow to the cores to perform multiple tasks of a process at the same time. Many bioinformatic programs use multiple cores/threads since it improves the speed of processing, however, it also increases the required RAM. Take as example, the workflow below, instead of waiting until the green task being performed to do the grey tasks, a multi-thread/core process can use core/threads to perform each task at the same time.
Useful commands…
ssh
- ssh [user@]hostname
- Is a program for logging/execute into a remote machine. It provides secure encrypted communications.
- It always requires a hostname, and it usually requires a username and a password.
htop
- htop
- Provides a dynamic real-time summary information of processes or threads currently being managed by the server. It is useful to know the available resources like CPUs and RAM.
free
- free
- Display amount of free and used memory in the system
screen
- screen; program X; Ctrl+a+d; program Y
- It allows you to execute the program X and let it run while you execute the Y program or do something else in the terminal.
- screen -r
The -r option allows you to return to the last screen.
Ctrl+a+d
- screen; program X; Ctrl+a+d; program Y
- It detaches you from the current screen
exit
- exit
- It allows you to close the screen windows or to log out of the server.
Ctrl + z
- python; Ctrl + z; cp ../aguacate.txt .; fg;
- It allows you to save the session of a program while you return to the terminal. If you are in python and you need to copy a file to the current directory, you can use Ctrl + z to go to the terminal without loosing the python terminal
- You can use it as many times as you wish, in other words, have multiple programs in execution
- fg will return you to the python session
fg
- python; Ctrl + z; cp ../aguacate.txt .; fg;
- It allows you to return to the programs in execution
scp
- scp FilePathOrigin FilePathDestiny; scp File user@host/:Path; scp user@host/:Path .
- It allows you to copy files from an origin directory to destiny directory. Both directories can be hosted in the same server/computer. However, the second command copies a file in the working directory to a server; the second one is the opposite.
ps
- ps
- list your current processes
pkill
- pkill ID#
- Kills the process with the ID ID#. You can check the IDs of your processes with ps.
date: Prints the date
whoami: Prints your username
hostname: prints the server/host name